Input data:
jpHMM result:
Sequence #1: D00330
This sequence is related to genotype(s): B C
| Fragment Start Position |
Uncertainty Region Start - End |
Breakpoint Interval Start - End |
Fragment End Position |
Fragment Genotype |
| Position in the original sequence [pred_recombination], [recombination_incl_UR_and_BPI], [UR_and_BPI] | ||||
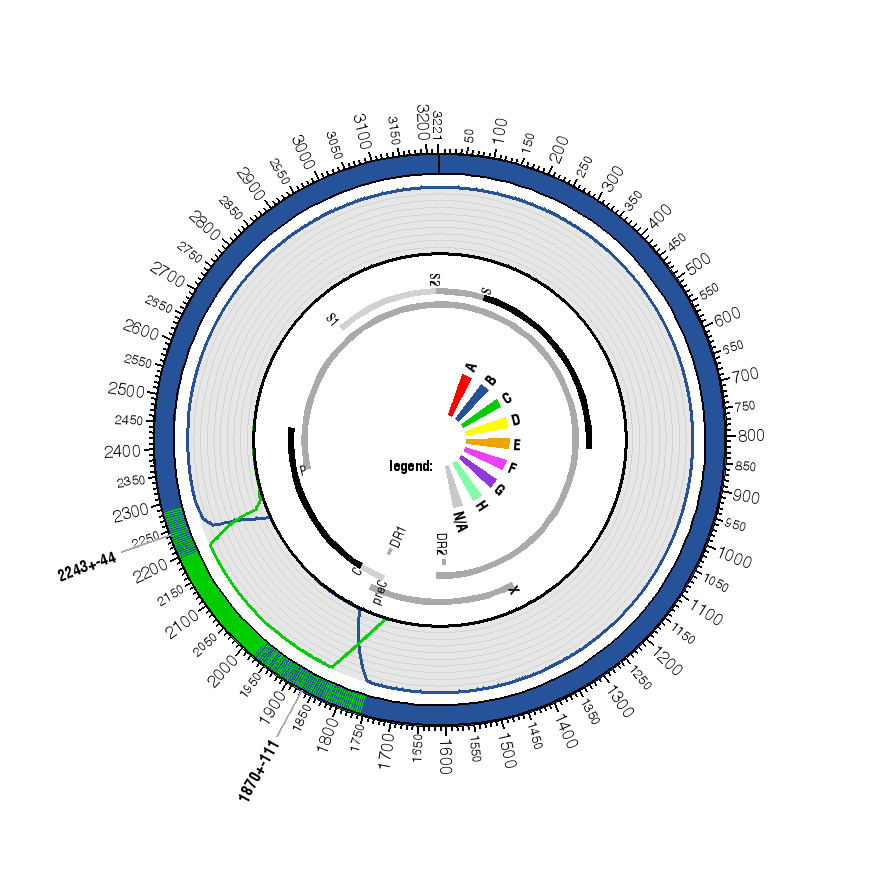
| 1 | - | 1759 - 1981 | 1837 | B |
| 1838 | - | 2199 - 2287 | 2232 | C |
| 2233 | - | - | 3215 | B |
| Position based on reference genome AM282986 numbering [pred_recombination] [recombination_incl_UR_and_BPI] [UR_and_BPI] | ||||
| 1 | - | 1759 - 1981 | 1837 | B |
| 1838 | - | 2199 - 2287 | 2232 | C |
| 2233 | - | - | 3221 | B |
| Genome map
(with position numbers given relative to the HBV reference genome
AM282986; the sequence has an offset of 0 to the reference origin (see pairwise alignment to reference sequence)): | ||||
 | ||||
Note:
| ||||
(Numbers x and y given in the output (">AM282986 x y") indicate the start (x) and end (y) position in the reference genome. Negative numbers x indicate an offset of x to the reference origin.)
Back to the jpHMM-HBV interface
Questions or comments? Email contact
Copyright © 2005-2006 Dept. of Bioinformatics (IMG)