Input data:
jpHMM result:
Sequence #1: >BF1_AX032749_EU446022
This sequence is related to subtype(s): B F1
| Fragment Start Position |
Uncertainty Region Start - End |
Breakpoint Interval Start - End |
Fragment End Position |
Fragment Subtype |
| Position in the original sequence [pred_recombination], [recombination_incl_UR_and_BPI], [UR_and_BPI] | ||||
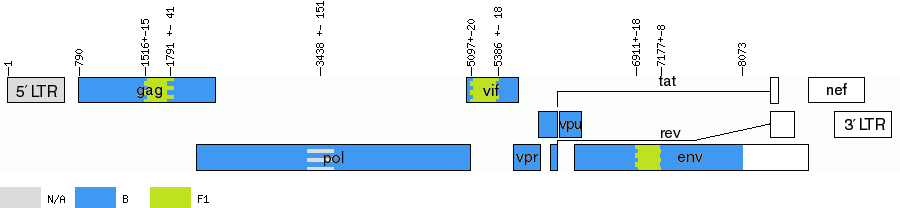
| 1 | - | - | 789 | N/A |
| 790 | - | 1501 - 1530 | 1530 | B |
| 1531 | - | 1750 - 1832 | 1808 | F1 |
| 1809 | 3287 - 3589 | 5077 - 5116 | 5095 | B |
| 5096 | - | 5368 - 5404 | 5397 | F1 |
| 5398 | - | 6883 - 6918 | 6899 | B |
| 6900 | - | 7153 - 7171 | 7165 | F1 |
| 7166 | - | - | 8060 | B |
| Position based on HXB2 numbering [pred_recombination] [recombination_incl_UR_and_BPI] [UR_and_BPI] | ||||
| 1 | - | - | 789 | N/A |
| 790 | - | 1501 - 1530 | 1530 | B |
| 1531 | - | 1750 - 1832 | 1808 | F1 |
| 1809 | 3287 - 3589 | 5077 - 5116 | 5095 | B |
| 5096 | - | 5368 - 5404 | 5397 | F1 |
| 5398 | - | 6893 - 6928 | 6909 | B |
| 6910 | - | 7169 - 7184 | 7178 | F1 |
| 7179 | - | - | 8073 | B |
| Genome map (based on HXB2 numbering) | ||||
 | ||||
Note:
| ||||
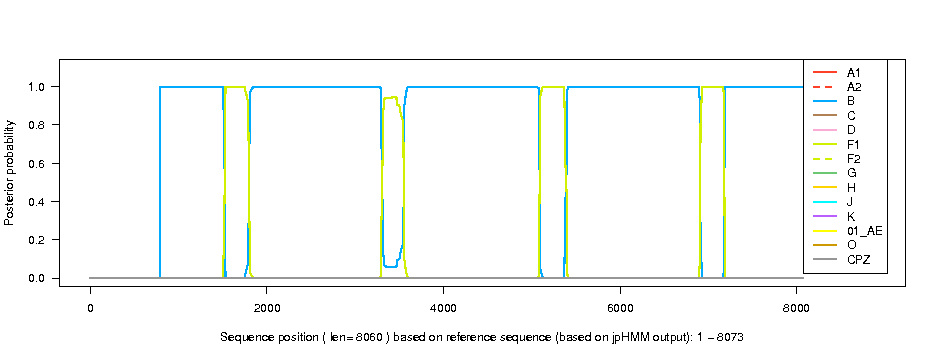
| Posterior probabilities of the subtypes (based on HXB2 numbering) | ||||
 | ||||
Back to the jpHMM-HIV interface
Questions or comments? Email contact
Copyright © 2005-2006 Dept. of Bioinformatics (IMG)